Note
Go to the end to download the full example code.
Sparse time artifact removal on simulated data#
This example is similar to test_nt_star.m in Noisetools. Results are equivalent (within numerical precision) to the Matlab code.
import matplotlib.pyplot as plt
import numpy as np
from meegkit import star
from meegkit.utils import demean, normcol
rng = np.random.default_rng(9)
Create simulated data#
Simulated data consist of N channels, 1 sinusoidal target, N-3 noise sources, with temporally local artifacts on each channel.
# Create simulated data
nchans = 10
n_samples = 1000
f = 2
target = np.sin(np.arange(n_samples) / n_samples * 2 * np.pi * f)
target = target[:, np.newaxis]
noise = rng.standard_normal((n_samples, nchans - 3))
# Create artifact signal
SNR = np.sqrt(1)
x0 = normcol(np.dot(noise, rng.standard_normal((noise.shape[1], nchans)))) + \
SNR * target * rng.standard_normal((1, nchans))
x0 = demean(x0)
artifact = np.zeros(x0.shape)
for k in np.arange(nchans):
artifact[k * 100 + np.arange(20), k] = 1
x = x0 + 10 * artifact
# This is to compare with matlab numerically
# from scipy.io import loadmat
# mat = loadmat('/Users/nicolas/Toolboxes/NoiseTools/TEST/X.mat')
# x = mat['x']
# x0 = mat['x0']
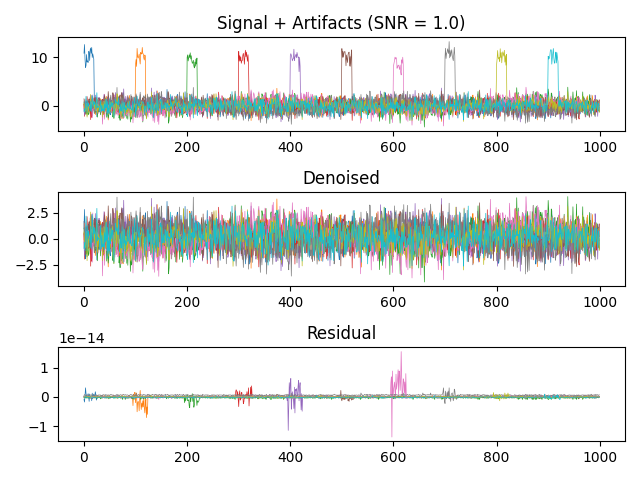
Apply STAR#
y, w, _ = star.star(x, 2)
proportion artifact free: 0.70
proportion artifact free: 0.70
proportion artifact free: 0.70
depth: 1
fixed channels: 10
fixed samples: 299
ratio: 1.01
power ratio: 0.40
Plot results#
f, (ax1, ax2, ax3) = plt.subplots(3, 1)
ax1.plot(x, lw=.5)
ax1.set_title(f"Signal + Artifacts (SNR = {SNR})")
ax2.plot(y, lw=.5)
ax2.set_title("Denoised")
ax3.plot(demean(y) - x0, lw=.5)
ax3.set_title("Residual")
f.set_tight_layout(True)
plt.show()
Total running time of the script: (0 minutes 0.301 seconds)

