Note
Go to the end to download the full example code.
Task-related component analysis for SSVEP detection#
Sample code for the task-related component analysis (TRCA)-based steady -state visual evoked potential (SSVEP) detection method [1]. The filter bank analysis can also be combined to the TRCA-based algorithm [2] [3].
This code is based on the Matlab implementation from: mnakanishi/TRCA-SSVEP
Uses meegkit.trca.TRCA().
References#
# Authors: Giuseppe Ferraro <giuseppe.ferraro@isae-supaero.fr>
# Nicolas Barascud <nicolas.barascud@gmail.com>
import os
import time
import matplotlib.pyplot as plt
import numpy as np
import scipy.io
from meegkit.trca import TRCA
from meegkit.utils.trca import itr, normfit, round_half_up
t = time.time()
Parameters#
dur_gaze = 0.5 # data length for target identification [s]
delay = 0.13 # visual latency being considered in the analysis [s]
n_bands = 5 # number of sub-bands in filter bank analysis
is_ensemble = True # True = ensemble TRCA method; False = TRCA method
alpha_ci = 0.05 # 100*(1-alpha_ci): confidence interval for accuracy
sfreq = 250 # sampling rate [Hz]
dur_shift = 0.5 # duration for gaze shifting [s]
list_freqs = np.array(
[[x + 8.0 for x in range(8)],
[x + 8.2 for x in range(8)],
[x + 8.4 for x in range(8)],
[x + 8.6 for x in range(8)],
[x + 8.8 for x in range(8)]]).T # list of stimulus frequencies
n_targets = list_freqs.size # The number of stimuli
# Useful variables (no need to modify)
dur_gaze_s = round_half_up(dur_gaze * sfreq) # data length [samples]
delay_s = round_half_up(delay * sfreq) # visual latency [samples]
dur_sel_s = dur_gaze + dur_shift # selection time [s]
ci = 100 * (1 - alpha_ci) # confidence interval
Load data#
path = os.path.join("..", "tests", "data", "trcadata.mat")
eeg = scipy.io.loadmat(path)["eeg"]
n_trials, n_chans, n_samples, n_blocks = eeg.shape
# Convert dummy Matlab format to (sample, channels, trials) and construct
# vector of labels
eeg = np.reshape(eeg.transpose([2, 1, 3, 0]),
(n_samples, n_chans, n_trials * n_blocks))
labels = np.array([x for x in range(n_targets)] * n_blocks)
crop_data = np.arange(delay_s, delay_s + dur_gaze_s)
eeg = eeg[crop_data]
TRCA classification#
Estimate classification performance with a Leave-One-Block-Out cross-validation approach.
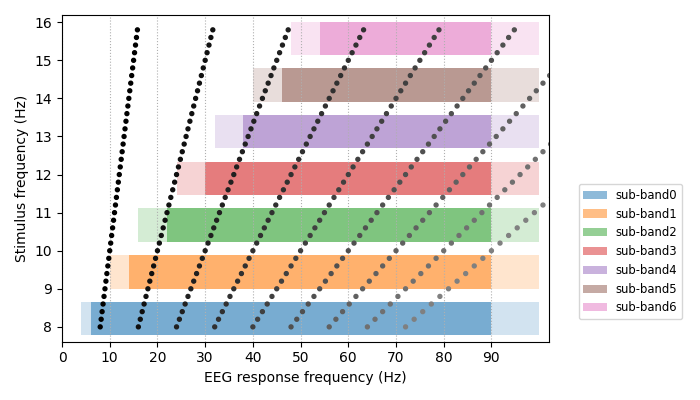
To get a sense of the filterbank specification in relation to the stimuli we can plot the individual filterbank sub-bands as well as the target frequencies (with their expected harmonics in the EEG spectrum). We use the filterbank specification described in [2].
filterbank = [[(6, 90), (4, 100)], # passband, stopband freqs [(Wp), (Ws)]
[(14, 90), (10, 100)],
[(22, 90), (16, 100)],
[(30, 90), (24, 100)],
[(38, 90), (32, 100)],
[(46, 90), (40, 100)],
[(54, 90), (48, 100)]]
f, ax = plt.subplots(1, figsize=(7, 4))
for i, _band in enumerate(filterbank):
ax.axvspan(ymin=i / len(filterbank) + .02,
ymax=(i + 1) / len(filterbank) - .02,
xmin=filterbank[i][1][0], xmax=filterbank[i][1][1],
alpha=0.2, facecolor=f"C{i}")
ax.axvspan(ymin=i / len(filterbank) + .02,
ymax=(i + 1) / len(filterbank) - .02,
xmin=filterbank[i][0][0], xmax=filterbank[i][0][1],
alpha=0.5, label=f"sub-band{i}", facecolor=f"C{i}")
for f in list_freqs.flat:
colors = np.ones((9, 4))
colors[:, :3] = np.linspace(0, .5, 9)[:, None]
ax.scatter(f * np.arange(1, 10), [f] * 9, c=colors, s=8, zorder=100)
ax.set_ylabel("Stimulus frequency (Hz)")
ax.set_xlabel("EEG response frequency (Hz)")
ax.set_xlim([0, 102])
ax.set_xticks(np.arange(0, 100, 10))
ax.grid(True, ls=":", axis="x")
ax.legend(bbox_to_anchor=(1.05, .5), fontsize="small")
plt.tight_layout()
plt.show()
Now perform the TRCA-based SSVEP detection algorithm
trca = TRCA(sfreq, filterbank, is_ensemble)
print("Results of the ensemble TRCA-based method:\n")
accs = np.zeros(n_blocks)
itrs = np.zeros(n_blocks)
for i in range(n_blocks):
# Select all folds except one for training
traindata = np.concatenate(
(eeg[..., :i * n_trials],
eeg[..., (i + 1) * n_trials:]), 2)
y_train = np.concatenate(
(labels[:i * n_trials], labels[(i + 1) * n_trials:]), 0)
# Construction of the spatial filter and the reference signals
trca.fit(traindata, y_train)
# Test stage
testdata = eeg[..., i * n_trials:(i + 1) * n_trials]
y_test = labels[i * n_trials:(i + 1) * n_trials]
estimated = trca.predict(testdata)
# Evaluation of the performance for this fold (accuracy and ITR)
is_correct = estimated == y_test
accs[i] = np.mean(is_correct) * 100
itrs[i] = itr(n_targets, np.mean(is_correct), dur_sel_s)
print(f"Block {i}: accuracy = {accs[i]:.1f}, \tITR = {itrs[i]:.1f}")
# Mean accuracy and ITR computation
mu, _, muci, _ = normfit(accs, alpha_ci)
print(f"\nMean accuracy = {mu:.1f}%\t({ci:.0f}% CI: {muci[0]:.1f}-{muci[1]:.1f}%)") # noqa
mu, _, muci, _ = normfit(itrs, alpha_ci)
print(f"Mean ITR = {mu:.1f}\t({ci:.0f}% CI: {muci[0]:.1f}-{muci[1]:.1f})")
if is_ensemble:
ensemble = "ensemble TRCA-based method"
else:
ensemble = "TRCA-based method"
print(f"\nElapsed time: {time.time()-t:.1f} seconds")
Results of the ensemble TRCA-based method:
Block 0: accuracy = 97.5, ITR = 301.3
Block 1: accuracy = 100.0, ITR = 319.3
Block 2: accuracy = 95.0, ITR = 286.3
Block 3: accuracy = 95.0, ITR = 286.3
Block 4: accuracy = 95.0, ITR = 286.3
Block 5: accuracy = 100.0, ITR = 319.3
Mean accuracy = 97.1% (95% CI: 97.0-97.1%)
Mean ITR = 299.8 (95% CI: 299.4-300.2)
Elapsed time: 11.7 seconds
Total running time of the script: (0 minutes 11.670 seconds)

